I would like to defend the use of paraphyletic groups in scientific discourse and literature. Paraphyletic groups can be well-defined in terms of monophyletic units (as relative complements), and defining paraphyletic groups in terms of monophyletic groups is preferable to treating them as invalid.
Let me start with a story. Wednesday evening (May 26th) I checked my Twitter feed, and saw a number of tweets from Jonathan Eisen (
phylogenomics), who was at the ASM meeting.

Jonathan is in the department of Ecology and Evolution at UC Davis, the author of a popular
textbook on Evolution and a frequent blogger ("
Tree of Life"). For those of you not used to reading Twitter feeds, note that the most recent tweets are at the top.
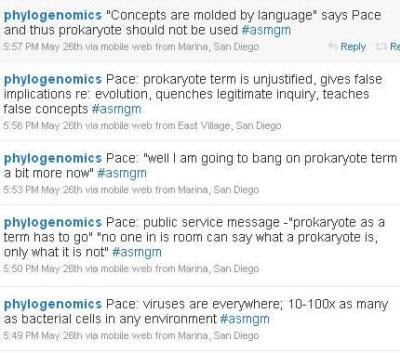

I know both Norm Pace and Jonathan Eisen. Thanks to Norm's personal style and Jonathan's excellent selection of quotes, reading this was like being in the room with Norm. I love hearing him talk. However, I do not entirely agree with him. I have spent my life studying gene expression in eukaryotes, and my perspective is that the differences between eukaryotes and other species ("prokaryotes") are fundamental. In prokaryotes, coupled transcription and translation (which is impossible when there is a nucleus) allows the widespread use of polycistronic mRNAs, which allow operons, which in turn contribute to many important features, including the ease with which biologically useful bits of genetic information can be horizontally transferred. The argument, repeated here by Norm Pace, that "no one can say what a prokaryote is, only what it is not" was addressed by
Martin and Koonin, who proposed a "positive definition of prokaryotes" based on coupled transcription and translation. This, however, is not the point. The point is that the nucleus is a derived feature and prokaryotes are a
paraphyletic group, meaning that the last common ancestor of all prokaryotes has descendants that are not prokaryotes. Nevertheless, the group is well-defined (as all life other than eukaryotes) and useful, so I commented:

A bit later, I commented again.
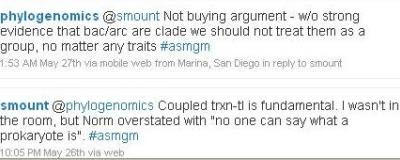
Prokaryotes are a paraphyletic group. That means that the last common ancestor of all prokaryotes has eukaryotic descendants. Most taxonomists today prefer not to talk about paraphyletic groups at all, but to speak only of monophyletic groups, or clades (which consist entirely of species with a common ancestor). However, there are many paraphyletic groups that "make sense" and are commonly used. Examples include prokaryotes, fish, reptiles and dicots.
My point is that defining a paraphyletic group as the
relative complement of one clade with respect to another makes it well-defined, and such a definition more closely suits what people have in mind.

In the hypothetical example shown here, most taxonomists would want to list "natural taxa" (by which they would mean monophyletic groups, or clades), and would say something like "Q, R and S are slithy." To say "G other than C are slithy" is more compact because it makes reference to fewer taxa. To say "P are slithy" is exactly the same, and is the most compact way of making the statement, but requires reference to a paraphyletic group.
To pursue this further, I asked my colleagues what they thought:
My dear friends in systematics,
I have a question about systematics that I would like your opinion on. It seems a sufficiently central question that I suspect you have already formed an opinion. The issue is a practical one, regarding how biologists should use terms. It is also philosophical (but in the rigorous sense, relating to the idea that without a proper philosophical basis one cannot do science at all).
Consider a monophyletic group of organisms, G, and another phylogenetic group within it, C (for clade). Let us suppose that C is characterized by some fundamental innovation, such that organisms within this clade have a long list of features not found in the other species within G. Furthermore, species within G but not C share a long list of features that have been lost by all species in C. As a result, there is a need to talk about another grouping, W (for wrong), of those species within G but not C. There is no doubt about the phylogeny. C and G are monophyletic but W is not. Molecules and morphology agree. However, all species within W share many features lacking in all species within C, and this is true both morphologically and molecularly.
Is it ever right for a scientist to talk about W as a group?
You know the list (reptiles, fish, dicots, prokaryotes).
Back story.
This came up last night as an argument between Jonathan Eisen and myself, on Twitter. You can see most of it by looking at feeds for
phylogenomics, ongenetics and smount, but given the volatile and perspective-based nature of Twitter feeds I've pasted the relevant tweets into the attached word document (it reads from most recent to earliest so my might want to start at the bottom and work up). Jonathan is at the ASM meeting. He is a Twitter addict who has generated over 4500 tweets in the last year or so (a day with only 10 would be unusual for him). I find it useful and interesting to follow him. I am both ongenetics and smount (I didn't mean to switch but I changed computers and forgot to switch).
I find it useful to refer to prokaryotes (and to fish). Jonathan says "grouping together bacteria/archaea is inappropriate; I note in my evolution textbook we use "bacteria & archaea" a lot". Wouldn't it be simpler if he just used "prokaryote."? I'm looking for advice here.
Thanks,
Steve
I received a thoughtful reply from Chuck Delwiche:
Well, I'm basically with Jonathan on this, although I think I'm slightly more moderate. "Fish," "prokaryotes," "reptiles," "dicots," etc. are really form-classes -- they describe the appearance of the organism, but not its evolutionary relationships. Naming paraphyletic groups is somewhat less objectionable than naming grossly polyphyletic ones, so I don't object to naming the North American Drosophila in a way that ignores the Hawaiian species that are derived from within it (this an example of your C/G case). But it really is confusing to refer to prokaryotes. Although they have coupled transcription and translation, the are other aspects of DNA replication, transcription, and translation that show striking similarities between Archaea and Eukarya. If you talk about "prokaryotes" as if the term represented a lineage rather than a morphology then it tends to obscure both diversity within them similarities between Archaea and Eukarya.
The reason this is important is that hides the predictive value that a natural classification can provide. Within your group G there would be some taxa that are more closely related to C than others, and they will share properties with C despite the long branch and loss of characters you describe. If you treat "fish" as a group it is confusing that Teleosts have immune systems that more nearly resemble those of tetrapods than do those of lampreys or hagfish. I don't know anything about lung- or lobe-finned fish immunology, but I'll bet they are even more tetrapod-like than those of Teleosts. Much the same statements could be made for skeletal structure, tooth anatomy, ventilation mechanisms, and I don't know what all else.
This is why we Must Never Speak of Fish Again.
Chuck
This is pretty much what I expected him to say, but there are two things I'd like to note. The first is this "
Within your group G there would be some taxa that are more closely related to C than others, and they will share properties with C despite the long branch and loss of characters you describe. If you treat "fish" as a group it is confusing that Teleosts have immune systems that more nearly resemble those of tetrapods than do those of lampreys or hagfish." This is a very good point that anyone who refers to paraphyletic groups must bear in mind.
The second thing that struck me is that he wrote "
'Fish,' 'prokaryotes,' 'reptiles,' 'dicots,' etc. are really form-classes -- they describe the appearance of the organism, but not its evolutionary relationships" despite the fact that I had provided a rigorous definition in terms of evolutionary relationships. Defining "fish" as vertebrates other than tetrapods makes it something other than a form class, and also eliminates any confusion about whales. Defining prokaryotes as organisms other than eukaryotes makes saying "prokaryotes" synonymous with saying "bacteria and archaea." It strikes me that this is a natural group in the sense that if a new domain of life were to be discovered (perhaps on Mars, or deep within the earth) with coupled transcription and translation and no nucleus or mitochondrion, people would want to group it with bacterial and archaea, even if phylogenetic analysis showed that it shared a most recent common ancestor with the eukaryotic nucleus.
In summary, I fully support the definition of taxa as monophyletic groups, and I would like to see them used to more rigorously define paraphyletic groups. Scientists will continue to refer to paraphyletic groups, and for good reasons. When they do, it would be useful if those groups were understood to be the relative complements of monophyletic taxa rather than informal categories, form classes or sloppy and unscientific categories.
I will continue to speak of fish. When I do, I will be referring to vertebrates that are not tetrapods. While I respect and understand colleagues who will never speak of fish, they must understand that this group is well-defined in terms of groups they recognize. I hope that all scientists move towards a more precise taxonomic basis for the groups that they will continue to talk about.
----------------------------------------
I thank Jonathan Eisen, Chuck Delwiche and Charlie Mitter for their contributions to this post. It goes without saying that the opinions expressed are, however, mine. The complete email thread (with Delwiche and Mitter) is available
here.
----------------------------------------
Postscript (June 13).
Charlie Mitter forwarded Farris 1979 (Systematic Zoology 28:483-519), which describes the state of systematics at that time as a debate between pheneticists, phylogeneticists and evolutionists about the principles that should underlie a general reference system for biology. I believe that this debate has been fully resolved in favor of the phylogeneticists, and I am fully persuaded that the business of systematics is the definition of monophyletic groups. My points here are that 1) biologists sometimes have good reasons to refer to paraphyletic groups and 2) when they do, it is better, where possible, to understand those groups in terms of monophyletic groups. It is precisely because I agree with the arguments of Farris in favor of phylogenetics that I think that the paraphyletic groups to which scientists will inevitably refer should be defined in terms of phylogenetic taxa (clades) and not thought of as elemental taxonomic units.





